
Improving Anaerobic Digestion of Protein for Meat Processors
by Dan McKeaton, Chemist and R&D, Aquafix, Inc
Abstract
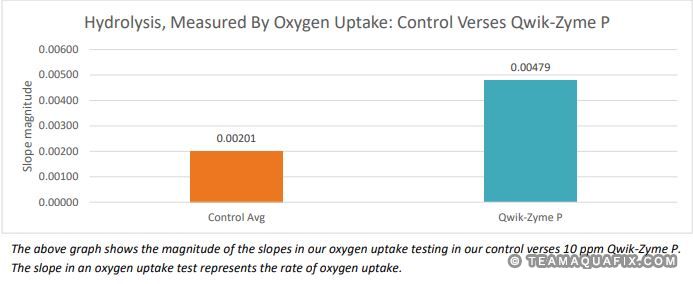
Qwik-Zyme P is a biocatalyst product designed to assist in the hydrolysis of complex proteins into free amino acids. In this study, we assessed the impacts of Qwik-Zyme P on poultry protein hydrolysis using oxygen uptake tests on samples collected from an anaerobic lagoon effluent, which enters into an aeration basin. An increase in oxygen uptake rate indicates improved aerobic degradation rates of the anaerobic effluent wastewater. As anaerobic systems tend to be inefficient at degrading proteins, we expected the anaerobic effluent to be high in protein content. This testing was run in two trials (5/19-5/20, 5/26-5/28). We observed an increase in oxygen uptake slope magnitude of 138% when we calculated the slope from the average data points in the two trials.
Methods
We performed a variety of different testing methods prior to running this final analysis. The method included here represents the final method used. This testing was run in triplicate and repeated with two separate sample sets to confirm results.
Anaerobic Effluent Sample
Anaerobic effluent grab samples were collected for this analysis. As anaerobic systems often have difficulty degrading proteins, it was believed that the anaerobic effluent from a poultry facility will be high in protein content. Samples were collected and shipped on ice and stored in the fridge. Fridge storage time (within one week) did not appear to make a difference in our analysis for the anaerobic effluent. All tests were run within one week of receiving the samples.
Bacterial Samples for analysis
In our analysis, we used our proprietary bacterial blend Yellow on 5/20/21, and our proprietary bacterial blend Purple on 5/26/21. Both blends are very similar and were believed to be interchangeable in this analysis. Bacterial samples were activated by adding 100 ppm of QLF Bio-P formula molasses. After adding the molasses, the Yellow/Purple blends were allowed to incubate at room temperature (23⁰C) overnight (approximately 18 hours). This was believed to allow for full degradation of the applied molasses to prevent molasses from interfering with oxygen uptake results.
Equipment
For oxygen uptake testing we used YSI-Multilab 4010-3W with 3xYSI ProOBOD probes. A 3-way adapter was required to attach all 3 probes to the meter. We mixed our reactors at 500 RPM using magnetic stir plates to ensure complete mixing.
Oxygen Uptake Procedure
For oxygen uptake testing, we prepared a blend of our activated bacterial product, anaerobic effluent, and dechlorinated tap water. This blend of 30 mL activated bacteria, 330 mL of anaerobic effluent, and 540 mL of dechlorinated tap water was well mixed, then distributed between 3 separate BOD bottles which were completely filled (300 mL per BOD bottle). Qwik-Zyme P was applied at 10 ppm to each of our Qwik-Zyme P reactors. No additive was applied to the controls. As previously stated, testing was run in triplicate. Probes and magnetic stir plates were rotated for each run to ensure consistency between probes. Reactors were prepared as follows:
5/19/21-5/20/21
Probe #32
Probe #70
Probe #72
5/25/21-5/28/21
Probe #32
Probe #70
Probe #72
The red cells in the table above indicate a sensor cap failure in probe #72. This required a 3rd run to complete our triplicate.
D.O. measurements were collected every 5 minutes throughout the testing. The 0-5 minutes time point is not factored into our analysis as this time point tends to very greatly between reactors and is heavily influenced by startup. Testing was run for a total of 240 minutes at room temperature.
Results
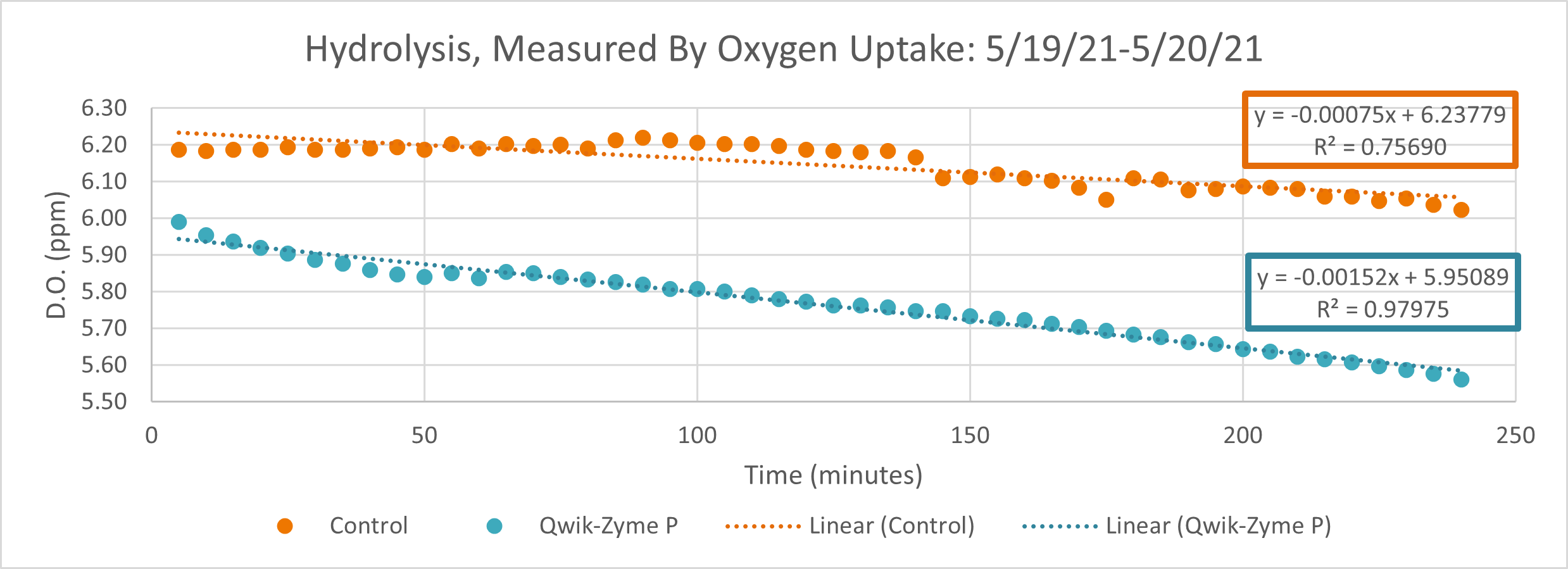
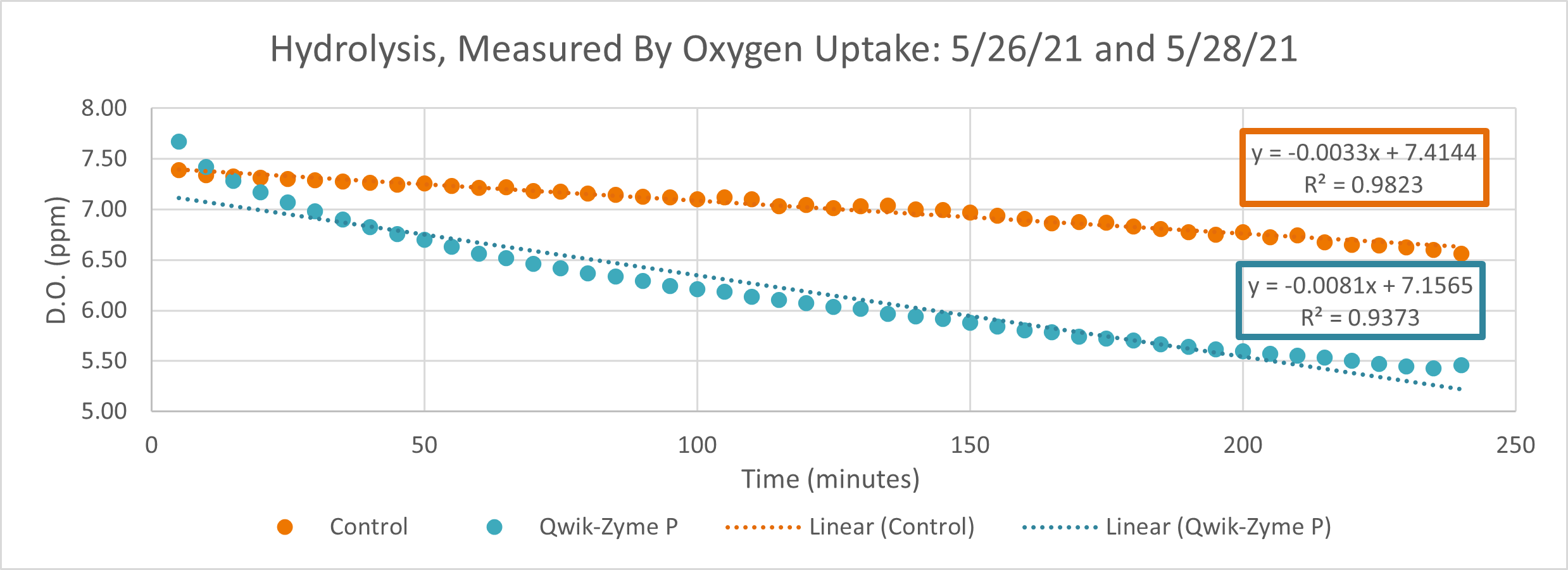
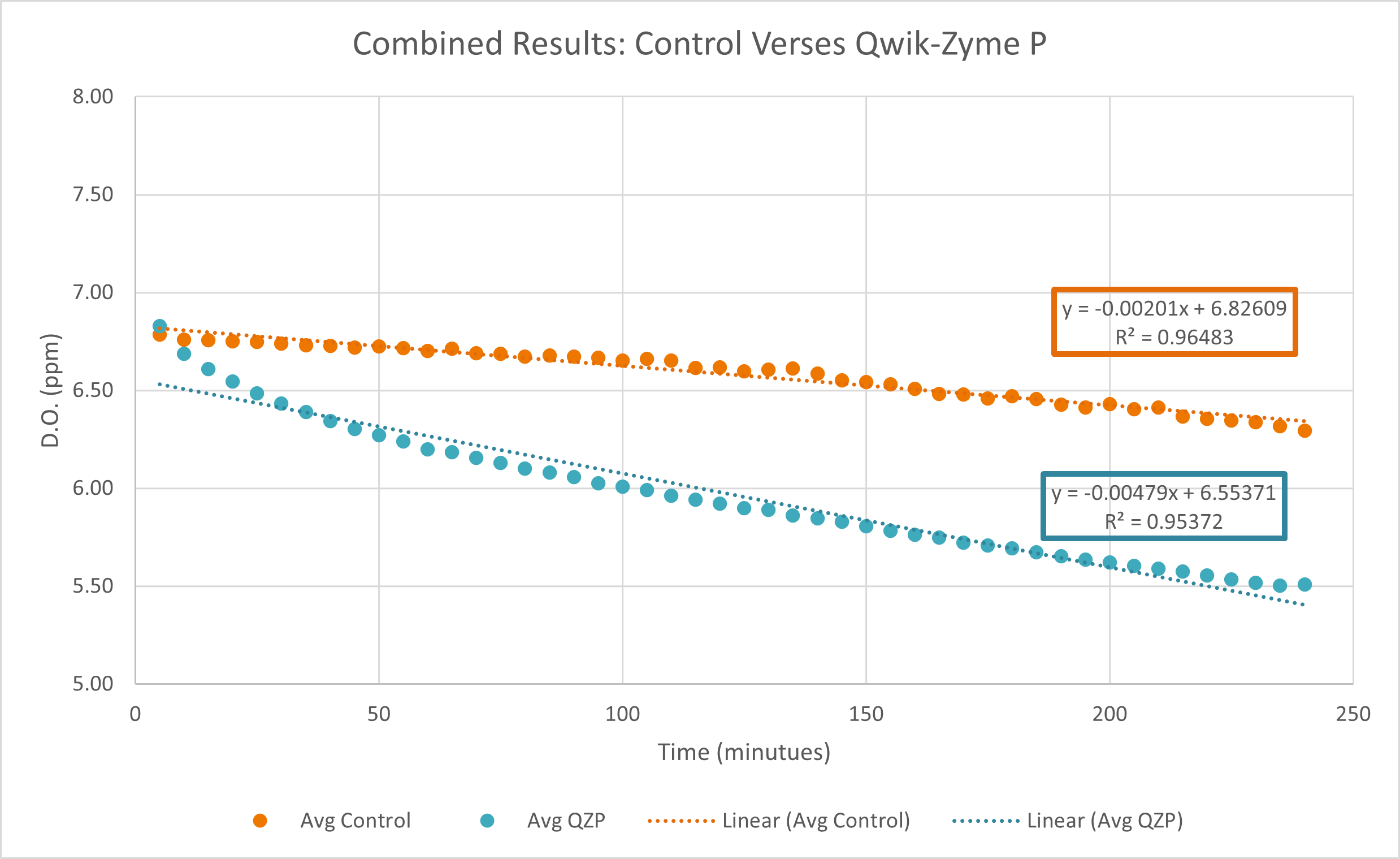
Conclusion
In this testing, the improvements in oxygen uptake rate confirmed that anaerobic effluent from this poultry facility was high in residual protein content, and that Qwik-Zyme P assisted in the degradation of this protein. The 138% increase in slope magnitude—observed when calculating the slope from the average of the datapoints between the two trials—is a relatively large change in oxygen uptake rates compared to what we’ve observed in other oxygen uptake trials. This testing suggests that anaerobic effluent from meat and poultry facilities is likely to be high in protein content. In comparing the control versus Qwik-Zyme P tested samples, results indicate that Qwik-Zyme P would be effective at assisting anaerobic poultry waste degradation in typical applications. It would be beneficial to run this testing in other similar facilities to confirm these results.
